Show code
### this is just the code that creates the "copy to clipboard" function in the code blocks
htmltools::tagList(
xaringanExtra::use_clipboard(
button_text = "<i class=\"fa fa-clone fa-2x\" style=\"color: #301e64\"></i>",
success_text = "<i class=\"fa fa-check fa-2x\" style=\"color: #90BE6D\"></i>",
error_text = "<i class=\"fa fa-times fa-2x\" style=\"color: #F94144\"></i>"
),
rmarkdown::html_dependency_font_awesome()
)Show code
as_tibble(list(x = 1,
y = 1)) -> tibDat
ggplot(data = tibDat,
aes(x = x, y = y)) +
geom_text(label = "Derangements #2",
size = 20,
colour = "red",
angle = 30,
lineheight = 1) +
xlab("") +
ylab("") +
theme_bw() +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.line = element_blank(),
axis.text = element_blank(),
axis.ticks = element_blank()) 
Updated to add contact me 11.ix.22
In my last post here, Scores from matching things I gave the background to the probabilities of achieving certain scores on matching tasks by chance alone to help explain the perhaps counter-intuitive finding that matching four or more things correctly is unlikely by chance alone at p < .05 regardless of the number of objects to be matched.
This just adds a bit more to that, mostly as plots and complements both that Rblog post and an “ordinary” blog post, Sometimes n=4 is enough.
What I wanted was to show how rapidly the probabilities of achieving any particular score stabilise to an asymptotic value as n increases. Here we are for n from 4 to 15 and scores from 4 to 10.
Show code
### create some functions (as in previous post)
all.derangements <- function(n){
cumprob <- prob <- number <- term <- score <- rev(0:n)
for (m in 1:n) {
i <- m+1
s <- n-m
term[i] <- ((-1)^(m))/(factorial(m))
}
term[1] <- 1
for (i in 0:n) {
s <- i+1
prob[s] <- (sum(term[1:s]))/factorial(n-i)
}
number <- factorial(n)*prob
for (s in 0:n) {
m <- n-s
i <- m+1
cumprob[i] <- sum(prob[1:i])
}
tmp <- cbind(n, score,number,prob,cumprob)
tmp
}
p.derange.score <- function(score,n){
if (score > n) stop("Score cannot be greater than n")
if (score == (n-1)) stop ("Score cannot be n-1")
cumprob <- prob <- term <- rev(0:n)
for (m in 1:n) {
i <- m+1
s <- n-m
term[i] <- ((-1)^(m))/(factorial(m))
}
term[1] <- 1
for (i in 0:n) {
s <- i+1
prob[s] <- (sum(term[1:s]))/factorial(n-i)
}
for (s in 0:n) {
m <- n-s
i <- m+1
cumprob[i] <- sum(prob[1:i])
}
cumprob[n+1-score]
}
### now let's go a bit further
### get all the possible scores for n from 4 to 30
lapply(4:30, FUN = all.derangements) -> tmpList
### I always forget this nice little bit of base R and I'm a bit surprised that there doesn't seem to be a nice tidyverse alternative
do.call(rbind.data.frame, tmpList) %>%
as_tibble() -> tmpTib
### this was just to produce some tables for my blog post at https://www.psyctc.org/psyctc/2022/07/23/sometimes-n4-is-enough/
# tmpTib %>%
# write_csv(file = "derangements.csv")
### ditto
# 1:14 %>%
# as_tibble() %>%
# rename(n = value) %>%
# mutate(PossibleWays = factorial(n),
# PossibleWays = prettyNum(PossibleWays, big.mark = ",")) %>%
# write_csv(file = "numbers.csv")
### but Vectorizing the function seemed cleaner so ...
Vectorize(FUN = all.derangements) -> All.derangements
All.derangements(4:14) -> tmpList
### back to the do.call() just for the tables
# do.call(rbind, tmpList) %>%
# as_tibble() %>%
# filter(score == 4) %>%
# select(n, number) %>%
# mutate(number = prettyNum(number, big.mark = ",")) %>%
# write_csv("correct.csv")
# do.call(rbind, tmpList) %>%
# as_tibble() %>%
# filter(score == 4) %>%
# mutate(totalPerms = factorial(n)) %>%
# select(-prob) %>%
# select(n, totalPerms, everything()) %>%
# write_csv("final.csv")
### OK, now for this Rblog post!
All.derangements(4:15) -> tmpList
do.call(rbind, tmpList) %>%
as_tibble() %>%
filter(score > 3 & score < 11) %>%
mutate(score = ordered(score,
levels = 4:10)) -> tmpTib
ggplot(data = tmpTib,
aes(x = n, y = cumprob, colour = score, group = score)) +
geom_point() +
geom_line() +
scale_x_continuous(breaks = 3:15,
minor_breaks = 3:15,
limits = c(3, 15)) +
ylab("p") +
theme_bw() 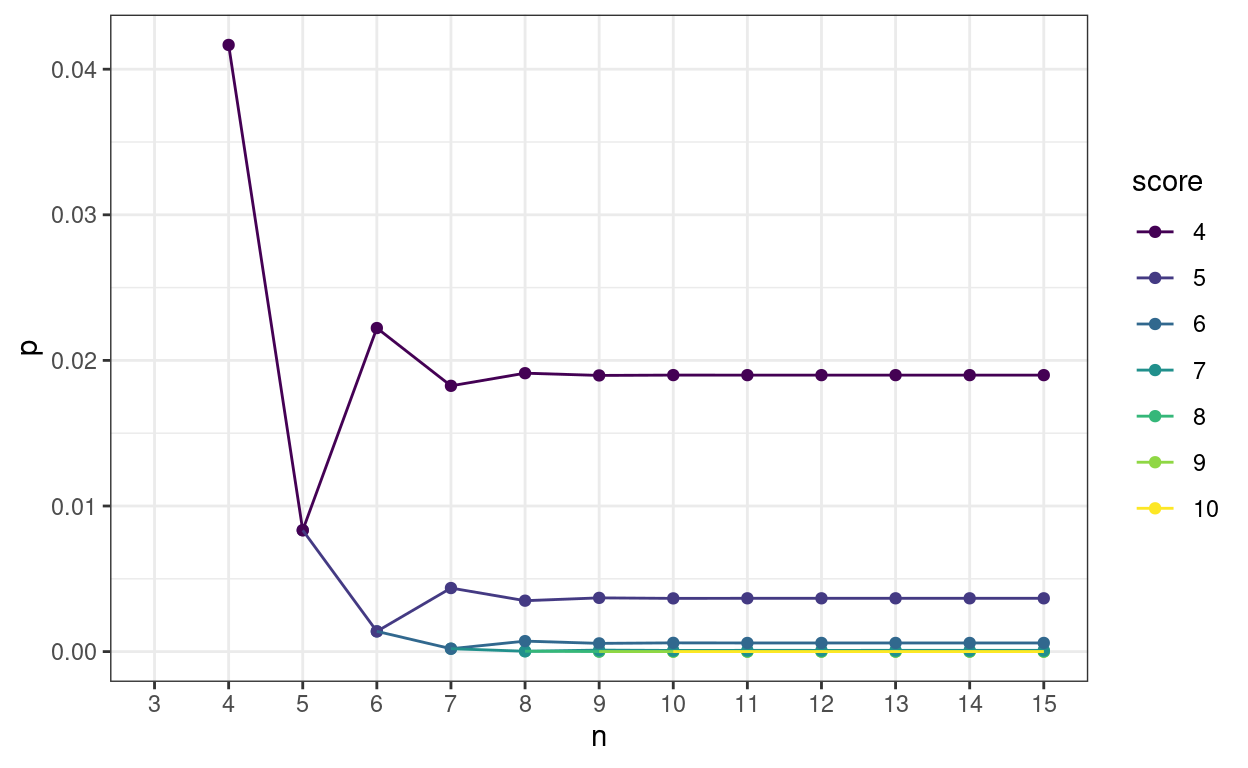
Here’s the same on a log10 y axis to separate the p values for the higher scores.
Show code
ggplot(data = tmpTib,
aes(x = n, y = cumprob, colour = score, group = score)) +
geom_point() +
geom_line() +
scale_x_continuous(breaks = 3:15,
minor_breaks = 3:15,
limits = c(3, 15)) +
scale_y_continuous(trans = "log10") +
ylab("p") +
theme_bw() 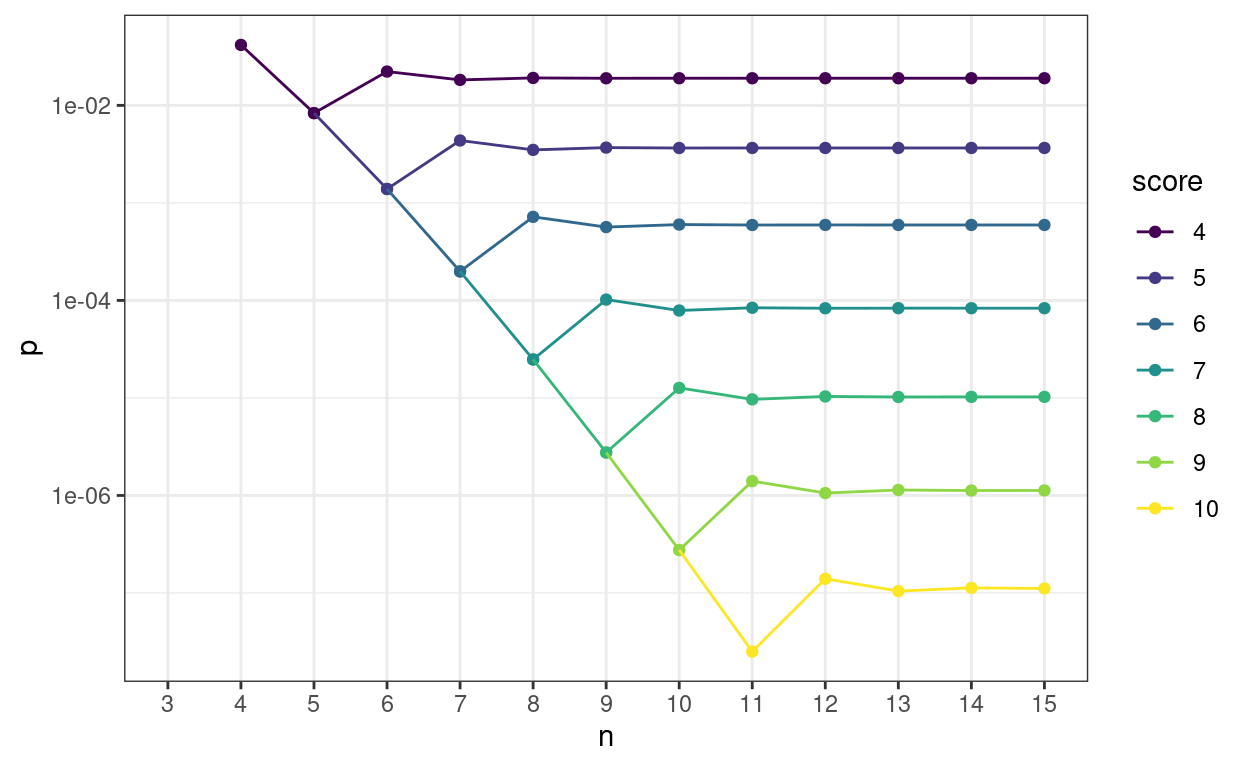
This next table shows how rapidly p values of real interest stabilise. The table is ordered by number of objects (n) within score. The column p is the probability of getting that score or better by chance alone, diffProb is the absolute change in that p value from the one for the previous n, diffPerc is the difference as a percentage of the previous p value. diffProbLT001 flags when the change in absolute p vaue is below .001 at which point I think in my realm any further precision is spurious. However, diffLT1pct flags when the change in p value is below 1% of the previous p value just in case someone wants that sort precise convergence.
Show code
All.derangements(4:20) -> tmpList
do.call(rbind, tmpList) %>%
as_tibble() %>%
filter(score > 3 & score < 11) -> tmpTib
### just working out how stable the p values get how soon
tmpTib %>%
arrange(score) %>%
group_by(score) %>%
mutate(diffProb = abs(cumprob - lag(cumprob)),
diffProbLT001 = if_else(diffProb < .001, "Y", "N"),
diffPerc = 100 * diffProb /lag(cumprob),
diffLT1pct = if_else(diffPerc < 1, "Y", "N")) %>%
ungroup() %>%
select(-c(number, prob)) %>%
rename(p = cumprob) %>%
select(score, everything()) %>%
as_hux() %>%
set_position("left") %>% # left align the whole table
set_bold(row = everywhere, col = everywhere) %>% # everything into bold
set_align(everywhere, everywhere, "center") %>% # everything centred
set_align(everywhere, 1:2, "right") %>% # but now right justify the first two columns
map_text_color(by_values("Y" = "green")) %>% # colour matches by text recognition
map_text_color(by_values("N" = "red"))| score | n | p | diffProb | diffProbLT001 | diffPerc | diffLT1pct |
|---|---|---|---|---|---|---|
| 4 | 4 | 0.0417 | ||||
| 4 | 5 | 0.00833 | 0.0333 | N | 80 | N |
| 4 | 6 | 0.0222 | 0.0139 | N | 167 | N |
| 4 | 7 | 0.0183 | 0.00397 | N | 17.9 | N |
| 4 | 8 | 0.0191 | 0.000868 | Y | 4.76 | N |
| 4 | 9 | 0.019 | 0.000154 | Y | 0.807 | Y |
| 4 | 10 | 0.019 | 2.31e-05 | Y | 0.122 | Y |
| 4 | 11 | 0.019 | 3.01e-06 | Y | 0.0158 | Y |
| 4 | 12 | 0.019 | 3.44e-07 | Y | 0.00181 | Y |
| 4 | 13 | 0.019 | 3.53e-08 | Y | 0.000186 | Y |
| 4 | 14 | 0.019 | 3.28e-09 | Y | 1.73e-05 | Y |
| 4 | 15 | 0.019 | 2.78e-10 | Y | 1.47e-06 | Y |
| 4 | 16 | 0.019 | 2.17e-11 | Y | 1.15e-07 | Y |
| 4 | 17 | 0.019 | 1.57e-12 | Y | 8.29e-09 | Y |
| 4 | 18 | 0.019 | 1.06e-13 | Y | 5.59e-10 | Y |
| 4 | 19 | 0.019 | 6.71e-15 | Y | 3.53e-11 | Y |
| 4 | 20 | 0.019 | 3.99e-16 | Y | 2.1e-12 | Y |
| 5 | 5 | 0.00833 | ||||
| 5 | 6 | 0.00139 | 0.00694 | N | 83.3 | N |
| 5 | 7 | 0.00437 | 0.00298 | N | 214 | N |
| 5 | 8 | 0.0035 | 0.000868 | Y | 19.9 | N |
| 5 | 9 | 0.00369 | 0.000193 | Y | 5.52 | N |
| 5 | 10 | 0.00366 | 3.47e-05 | Y | 0.941 | Y |
| 5 | 11 | 0.00366 | 5.26e-06 | Y | 0.144 | Y |
| 5 | 12 | 0.00366 | 6.89e-07 | Y | 0.0188 | Y |
| 5 | 13 | 0.00366 | 7.95e-08 | Y | 0.00217 | Y |
| 5 | 14 | 0.00366 | 8.2e-09 | Y | 0.000224 | Y |
| 5 | 15 | 0.00366 | 7.65e-10 | Y | 2.09e-05 | Y |
| 5 | 16 | 0.00366 | 6.52e-11 | Y | 1.78e-06 | Y |
| 5 | 17 | 0.00366 | 5.12e-12 | Y | 1.4e-07 | Y |
| 5 | 18 | 0.00366 | 3.72e-13 | Y | 1.02e-08 | Y |
| 5 | 19 | 0.00366 | 2.52e-14 | Y | 6.87e-10 | Y |
| 5 | 20 | 0.00366 | 1.59e-15 | Y | 4.35e-11 | Y |
| 6 | 6 | 0.00139 | ||||
| 6 | 7 | 0.000198 | 0.00119 | N | 85.7 | N |
| 6 | 8 | 0.000719 | 0.000521 | Y | 262 | N |
| 6 | 9 | 0.000565 | 0.000154 | Y | 21.5 | N |
| 6 | 10 | 0.0006 | 3.47e-05 | Y | 6.15 | N |
| 6 | 11 | 0.000593 | 6.31e-06 | Y | 1.05 | N |
| 6 | 12 | 0.000594 | 9.65e-07 | Y | 0.163 | Y |
| 6 | 13 | 0.000594 | 1.27e-07 | Y | 0.0214 | Y |
| 6 | 14 | 0.000594 | 1.48e-08 | Y | 0.00248 | Y |
| 6 | 15 | 0.000594 | 1.53e-09 | Y | 0.000258 | Y |
| 6 | 16 | 0.000594 | 1.44e-10 | Y | 2.42e-05 | Y |
| 6 | 17 | 0.000594 | 1.23e-11 | Y | 2.07e-06 | Y |
| 6 | 18 | 0.000594 | 9.67e-13 | Y | 1.63e-07 | Y |
| 6 | 19 | 0.000594 | 7.04e-14 | Y | 1.19e-08 | Y |
| 6 | 20 | 0.000594 | 4.78e-15 | Y | 8.04e-10 | Y |
| 7 | 7 | 0.000198 | ||||
| 7 | 8 | 2.48e-05 | 0.000174 | Y | 87.5 | N |
| 7 | 9 | 0.000102 | 7.72e-05 | Y | 311 | N |
| 7 | 10 | 7.88e-05 | 2.31e-05 | Y | 22.7 | N |
| 7 | 11 | 8.41e-05 | 5.26e-06 | Y | 6.68 | N |
| 7 | 12 | 8.31e-05 | 9.65e-07 | Y | 1.15 | N |
| 7 | 13 | 8.33e-05 | 1.48e-07 | Y | 0.179 | Y |
| 7 | 14 | 8.32e-05 | 1.97e-08 | Y | 0.0236 | Y |
| 7 | 15 | 8.32e-05 | 2.3e-09 | Y | 0.00276 | Y |
| 7 | 16 | 8.32e-05 | 2.39e-10 | Y | 0.000287 | Y |
| 7 | 17 | 8.32e-05 | 2.25e-11 | Y | 2.7e-05 | Y |
| 7 | 18 | 8.32e-05 | 1.93e-12 | Y | 2.32e-06 | Y |
| 7 | 19 | 8.32e-05 | 1.53e-13 | Y | 1.83e-07 | Y |
| 7 | 20 | 8.32e-05 | 1.12e-14 | Y | 1.34e-08 | Y |
| 8 | 8 | 2.48e-05 | ||||
| 8 | 9 | 2.76e-06 | 2.2e-05 | Y | 88.9 | N |
| 8 | 10 | 1.27e-05 | 9.92e-06 | Y | 360 | N |
| 8 | 11 | 9.67e-06 | 3.01e-06 | Y | 23.7 | N |
| 8 | 12 | 1.04e-05 | 6.89e-07 | Y | 7.12 | N |
| 8 | 13 | 1.02e-05 | 1.27e-07 | Y | 1.23 | N |
| 8 | 14 | 1.03e-05 | 1.97e-08 | Y | 0.192 | Y |
| 8 | 15 | 1.02e-05 | 2.62e-09 | Y | 0.0256 | Y |
| 8 | 16 | 1.02e-05 | 3.08e-10 | Y | 0.003 | Y |
| 8 | 17 | 1.02e-05 | 3.22e-11 | Y | 0.000314 | Y |
| 8 | 18 | 1.02e-05 | 3.04e-12 | Y | 2.96e-05 | Y |
| 8 | 19 | 1.02e-05 | 2.62e-13 | Y | 2.55e-06 | Y |
| 8 | 20 | 1.02e-05 | 2.07e-14 | Y | 2.02e-07 | Y |
| 9 | 9 | 2.76e-06 | ||||
| 9 | 10 | 2.76e-07 | 2.48e-06 | Y | 90 | N |
| 9 | 11 | 1.4e-06 | 1.13e-06 | Y | 409 | N |
| 9 | 12 | 1.06e-06 | 3.44e-07 | Y | 24.6 | N |
| 9 | 13 | 1.14e-06 | 7.95e-08 | Y | 7.51 | N |
| 9 | 14 | 1.12e-06 | 1.48e-08 | Y | 1.3 | N |
| 9 | 15 | 1.13e-06 | 2.3e-09 | Y | 0.204 | Y |
| 9 | 16 | 1.13e-06 | 3.08e-10 | Y | 0.0273 | Y |
| 9 | 17 | 1.13e-06 | 3.62e-11 | Y | 0.00322 | Y |
| 9 | 18 | 1.13e-06 | 3.8e-12 | Y | 0.000337 | Y |
| 9 | 19 | 1.13e-06 | 3.6e-13 | Y | 3.2e-05 | Y |
| 9 | 20 | 1.13e-06 | 3.11e-14 | Y | 2.76e-06 | Y |
| 10 | 10 | 2.76e-07 | ||||
| 10 | 11 | 2.51e-08 | 2.51e-07 | Y | 90.9 | N |
| 10 | 12 | 1.4e-07 | 1.15e-07 | Y | 458 | N |
| 10 | 13 | 1.05e-07 | 3.53e-08 | Y | 25.3 | N |
| 10 | 14 | 1.13e-07 | 8.2e-09 | Y | 7.85 | N |
| 10 | 15 | 1.11e-07 | 1.53e-09 | Y | 1.36 | N |
| 10 | 16 | 1.11e-07 | 2.39e-10 | Y | 0.215 | Y |
| 10 | 17 | 1.11e-07 | 3.22e-11 | Y | 0.0289 | Y |
| 10 | 18 | 1.11e-07 | 3.8e-12 | Y | 0.00341 | Y |
| 10 | 19 | 1.11e-07 | 4e-13 | Y | 0.000359 | Y |
| 10 | 20 | 1.11e-07 | 3.8e-14 | Y | 3.41e-05 | Y |
Do contact me if this interests you and if you might want to use the method with real data.
free web page counter